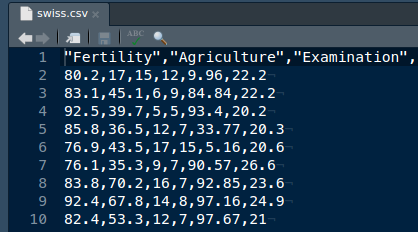
swiss Fertility Agriculture Examination Education Catholic
Courtelary 80.2 17.0 15 12 9.96
Delemont 83.1 45.1 6 9 84.84
Franches-Mnt 92.5 39.7 5 5 93.40
Moutier 85.8 36.5 12 7 33.77
Neuveville 76.9 43.5 17 15 5.16
Porrentruy 76.1 35.3 9 7 90.57
Broye 83.8 70.2 16 7 92.85
Glane 92.4 67.8 14 8 97.16
Gruyere 82.4 53.3 12 7 97.67
Sarine 82.9 45.2 16 13 91.38
Veveyse 87.1 64.5 14 6 98.61
Aigle 64.1 62.0 21 12 8.52
Aubonne 66.9 67.5 14 7 2.27
Avenches 68.9 60.7 19 12 4.43
Cossonay 61.7 69.3 22 5 2.82
Echallens 68.3 72.6 18 2 24.20
Grandson 71.7 34.0 17 8 3.30
Lausanne 55.7 19.4 26 28 12.11
La Vallee 54.3 15.2 31 20 2.15
Lavaux 65.1 73.0 19 9 2.84
Morges 65.5 59.8 22 10 5.23
Moudon 65.0 55.1 14 3 4.52
Nyone 56.6 50.9 22 12 15.14
Orbe 57.4 54.1 20 6 4.20
Oron 72.5 71.2 12 1 2.40
Payerne 74.2 58.1 14 8 5.23
Paysd'enhaut 72.0 63.5 6 3 2.56
Rolle 60.5 60.8 16 10 7.72
Vevey 58.3 26.8 25 19 18.46
Yverdon 65.4 49.5 15 8 6.10
Conthey 75.5 85.9 3 2 99.71
Entremont 69.3 84.9 7 6 99.68
Herens 77.3 89.7 5 2 100.00
Martigwy 70.5 78.2 12 6 98.96
Monthey 79.4 64.9 7 3 98.22
St Maurice 65.0 75.9 9 9 99.06
Sierre 92.2 84.6 3 3 99.46
Sion 79.3 63.1 13 13 96.83
Boudry 70.4 38.4 26 12 5.62
La Chauxdfnd 65.7 7.7 29 11 13.79
Le Locle 72.7 16.7 22 13 11.22
Neuchatel 64.4 17.6 35 32 16.92
Val de Ruz 77.6 37.6 15 7 4.97
ValdeTravers 67.6 18.7 25 7 8.65
V. De Geneve 35.0 1.2 37 53 42.34
Rive Droite 44.7 46.6 16 29 50.43
Rive Gauche 42.8 27.7 22 29 58.33
Infant.Mortality
Courtelary 22.2
Delemont 22.2
Franches-Mnt 20.2
Moutier 20.3
Neuveville 20.6
Porrentruy 26.6
Broye 23.6
Glane 24.9
Gruyere 21.0
Sarine 24.4
Veveyse 24.5
Aigle 16.5
Aubonne 19.1
Avenches 22.7
Cossonay 18.7
Echallens 21.2
Grandson 20.0
Lausanne 20.2
La Vallee 10.8
Lavaux 20.0
Morges 18.0
Moudon 22.4
Nyone 16.7
Orbe 15.3
Oron 21.0
Payerne 23.8
Paysd'enhaut 18.0
Rolle 16.3
Vevey 20.9
Yverdon 22.5
Conthey 15.1
Entremont 19.8
Herens 18.3
Martigwy 19.4
Monthey 20.2
St Maurice 17.8
Sierre 16.3
Sion 18.1
Boudry 20.3
La Chauxdfnd 20.5
Le Locle 18.9
Neuchatel 23.0
Val de Ruz 20.0
ValdeTravers 19.5
V. De Geneve 18.0
Rive Droite 18.2
Rive Gauche 19.3