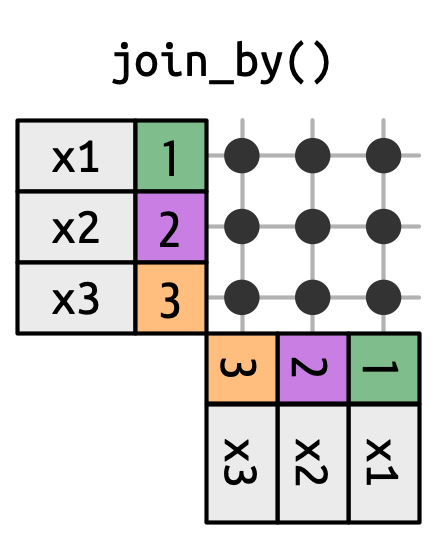
Joins for tables
dplyr
University of Luxembourg
Saturday, the 25th of April, 2026
Relational operations
Combining data from two tables
How do we refer to a particular observation?
Primary key
The column that uniquely identifies an observation
- Frequently an ID
- Can be introduced by
row_number()if no primary column exists bona fide observations
Composite key
Several columns that together identify an observation uniquely to serve as primary key.
Like patient and visit
Foreign key
A key referring to observations in another table
Your turn!
Which columns are keys?
- In
judgments
- In
datasauRus::datasaurus_dozen - In
swiss
Warning
Tibbles do not have row names! Frequently these are the primary keys!
Use as_tibble(data, rownames = "ID") for swiss
Solution
swiss: Province is primary key.
Once you took care of the rownames.
# A tibble: 47 × 7
Province Fertility Agriculture Examination Education Catholic
<chr> <dbl> <dbl> <int> <int> <dbl>
1 Courtelary 80.2 17 15 12 9.96
2 Delemont 83.1 45.1 6 9 84.8
3 Franches-Mnt 92.5 39.7 5 5 93.4
4 Moutier 85.8 36.5 12 7 33.8
5 Neuveville 76.9 43.5 17 15 5.16
6 Porrentruy 76.1 35.3 9 7 90.6
7 Broye 83.8 70.2 16 7 92.8
8 Glane 92.4 67.8 14 8 97.2
9 Gruyere 82.4 53.3 12 7 97.7
10 Sarine 82.9 45.2 16 13 91.4
# ℹ 37 more rows
# ℹ 1 more variable: Infant.Mortality <dbl>Datasaurus: composite key from all the variables
# A tibble: 5 × 3
dataset x y
<chr> <dbl> <dbl>
1 away 61.9 71.8
2 v_lines 89.5 46.6
3 dino 46.9 79.9
4 v_lines 69.5 16.9
5 dots 77.7 51.2Or a primary key from row numbers
# A tibble: 1,846 × 4
ID dataset x y
<chr> <chr> <dbl> <dbl>
1 1 dino 55.4 97.2
2 2 dino 51.5 96.0
3 3 dino 46.2 94.5
4 4 dino 42.8 91.4
5 5 dino 40.8 88.3
6 6 dino 38.7 84.9
7 7 dino 35.6 79.9
8 8 dino 33.1 77.6
9 9 dino 29.0 74.5
10 10 dino 26.2 71.4
# ℹ 1,836 more rowsPrimary key in judgments
Should be subject
# A tibble: 188 × 1
subject
<dbl>
1 2
2 1
3 3
4 4
5 7
6 6
7 5
8 9
9 16
10 13
# ℹ 178 more rowsMatching subjects between two tibbles
Additional data for some participants
- Coffee consumption
tribble(~student, ~coffee_shots,
21, 1,
23, 4,
1 211, 3,
28, 2) -> coffee_drinkers
coffee_drinkers- 1
- Not present in the initial data
# A tibble: 4 × 2
student coffee_shots
<dbl> <dbl>
1 21 1
2 23 4
3 211 3
4 28 2Smaller sample set
# A tibble: 187 × 5
subject condition gender mood_pre mood_post
<dbl> <chr> <chr> <dbl> <dbl>
1 2 control female 81 NA
2 1 stress female 59 42
3 3 stress female 22 60
4 4 stress female 53 68
5 7 control female 48 NA
6 6 stress female 73 73
7 5 control female NA NA
8 9 control male 100 NA
9 16 stress female 67 74
10 13 stress female 30 68
# ℹ 177 more rowsGoal: Add coffee consumption
This is made up data for demonstration purposes only.
Combining the two tables
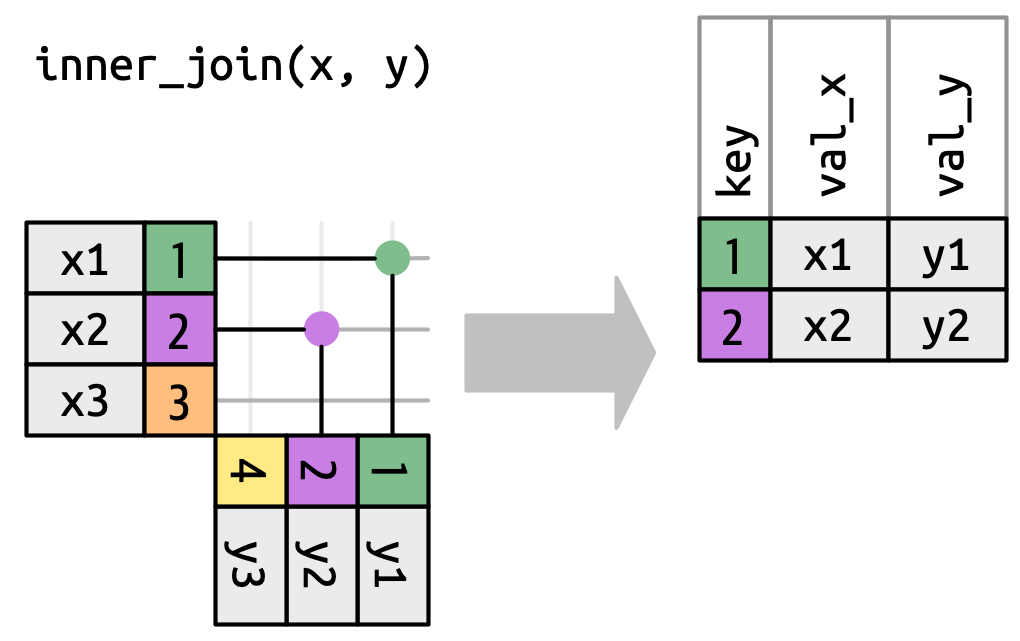
Inner join

Error! no common key.
Error in `inner_join()`:
! `by` must be supplied when `x` and `y` have no common variables.
ℹ Use `cross_join()` to perform a cross-join.Provide the correspondences
# A tibble: 3 × 6
student coffee_shots condition gender mood_pre mood_post
<dbl> <dbl> <chr> <chr> <dbl> <dbl>
1 21 1 control female 68 NA
2 23 4 control female 78 NA
3 28 2 stress male 53 68Initial tables are not preserved, but mutated
Mutating joins
Creating new tables through joins
- Key operations in data processing
inner_join()is the most strict join operationsleft_join()focuses on left tableright_join()focuses on right tablefull_join()joins both tables- Missing values can create massive matches
- left, right, full create missing values for unmatched rows
Join types
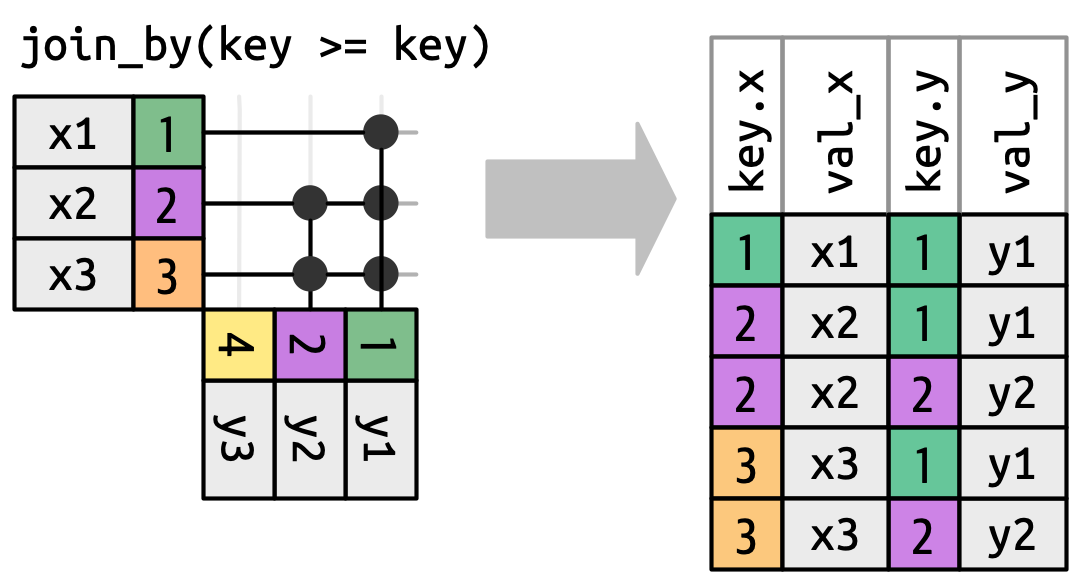
From the by = argument and specified in the join_by() helper:
- Equality joins (
==) - Inequality joins (
>=) - Rolling joins (
closest(a >= b)) - Overlap joins (
between() / within() / overlaps()) - Cross joins, returns Cartesian product
nrow(x) * nrow(y)
Mutating joins, overview
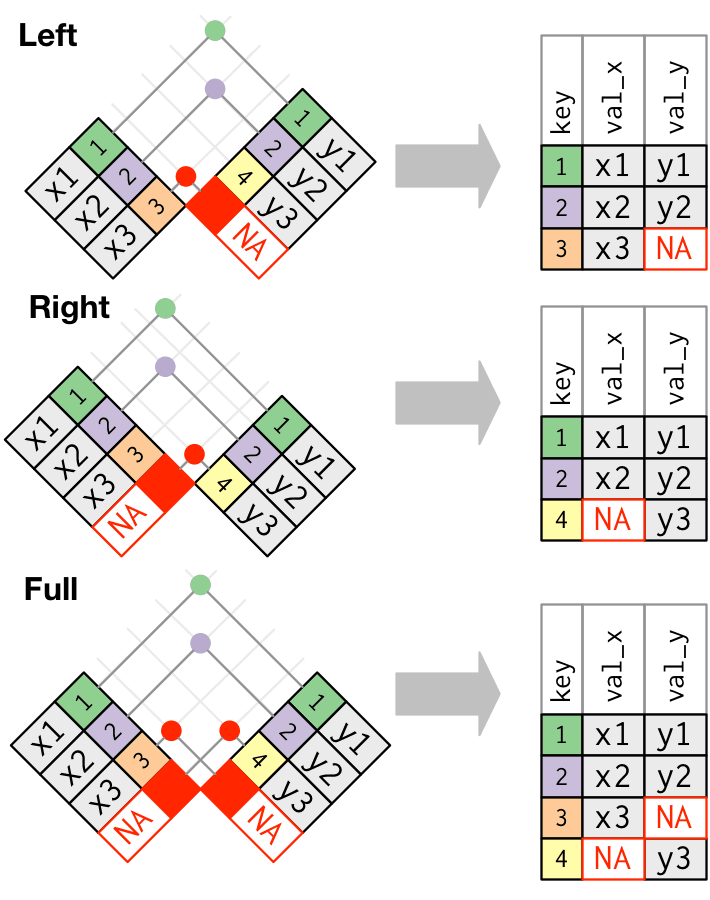
Left join
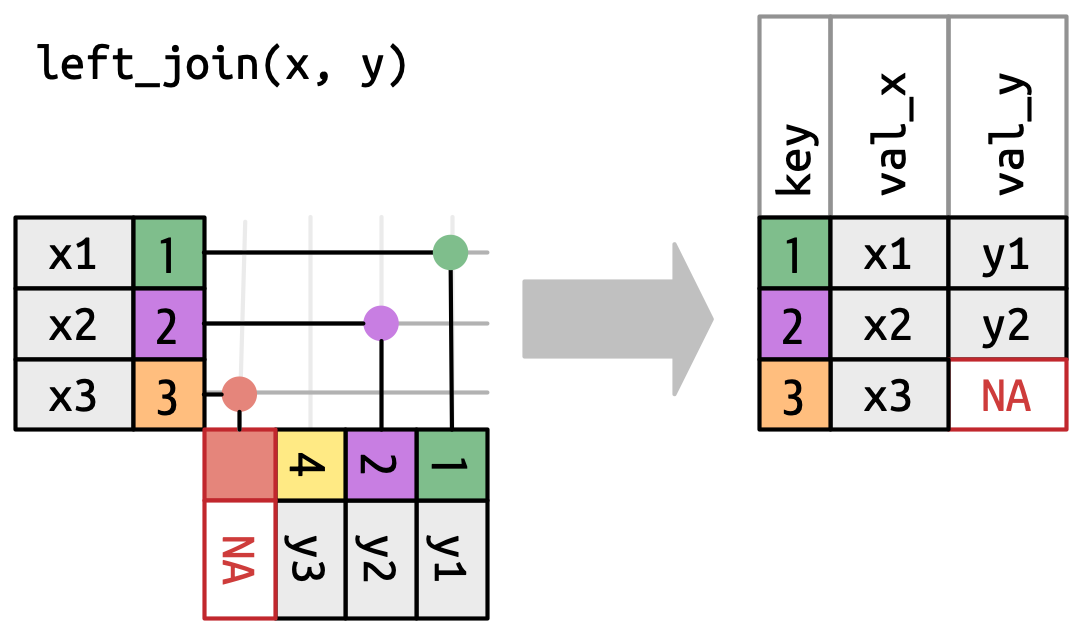
Left tibble keeps same numbers of rows.
Example with left_join()
# A tibble: 187 × 6
subject condition gender mood_pre mood_post coffee_shots
<dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 2 control female 81 NA NA
2 1 stress female 59 42 NA
3 3 stress female 22 60 NA
4 4 stress female 53 68 NA
5 7 control female 48 NA NA
6 6 stress female 73 73 NA
7 5 control female NA NA NA
8 9 control male 100 NA NA
9 16 stress female 67 74 NA
10 13 stress female 30 68 NA
# ℹ 177 more rowsThe missing subject 211 is unmatched and not returned
Right-join
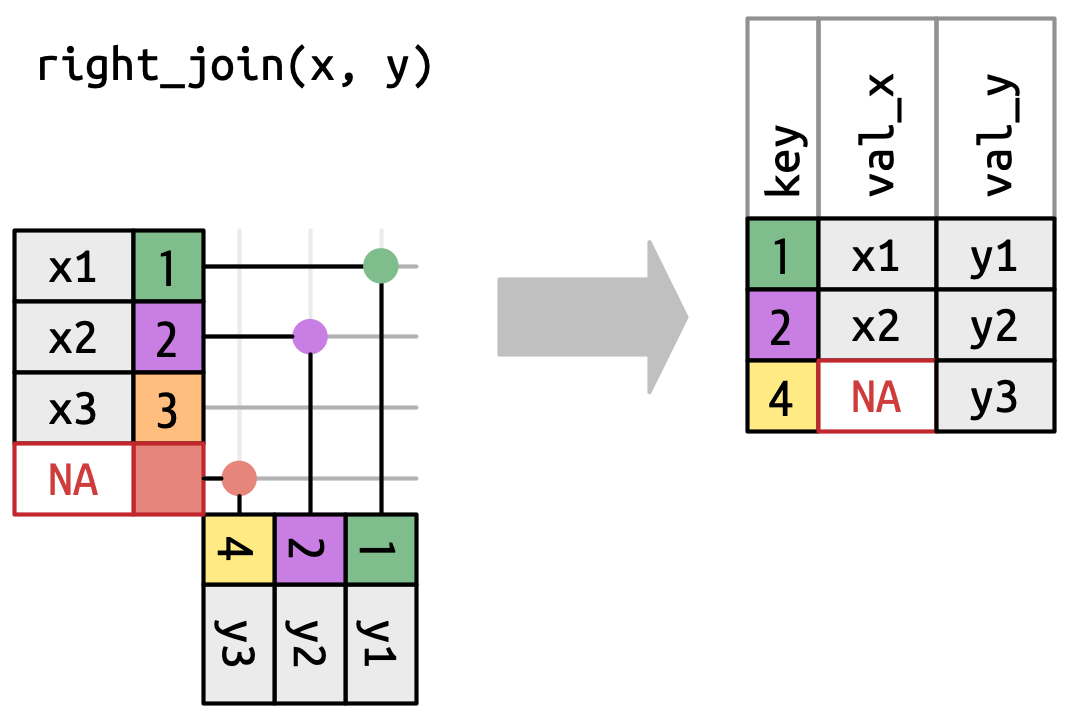
Right tibble matters
Example with right_join()
# A tibble: 4 × 6
subject condition gender mood_pre mood_post coffee_shots
<dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 23 control female 78 NA 4
2 21 control female 68 NA 1
3 28 stress male 53 68 2
4 211 <NA> <NA> NA NA 3The missing subject 211 is unmatched and returned with NA for left columns
Full-join
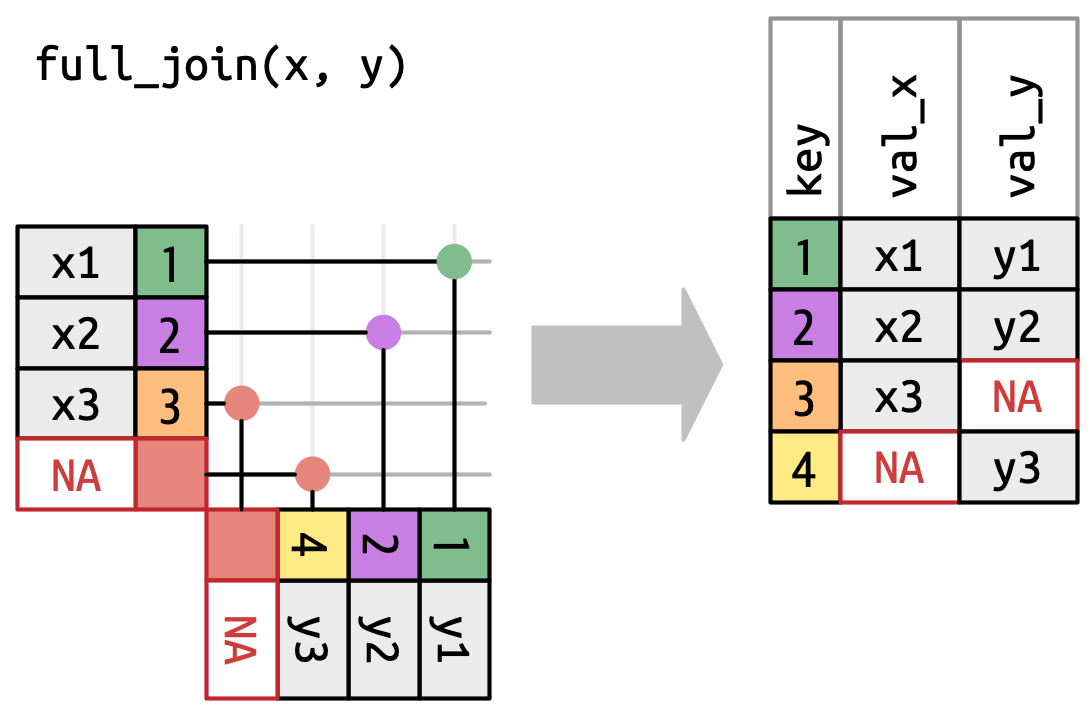
Number of rows = left + right
Example with full_join()
# A tibble: 188 × 6
subject condition gender mood_pre mood_post coffee_shots
<dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 2 control female 81 NA NA
2 1 stress female 59 42 NA
3 3 stress female 22 60 NA
4 4 stress female 53 68 NA
5 7 control female 48 NA NA
6 6 stress female 73 73 NA
7 5 control female NA NA NA
8 9 control male 100 NA NA
9 16 stress female 67 74 NA
10 13 stress female 30 68 NA
# ℹ 178 more rowsThe missing subject 211 is unmatched and returned with NAs
# A tibble: 4 × 6
subject condition gender mood_pre mood_post coffee_shots
<dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 23 control female 78 NA 4
2 21 control female 68 NA 1
3 28 stress male 53 68 2
4 211 <NA> <NA> NA NA 3Two tables - same column names
What if we have gender in both tables?
# A tibble: 4 × 3
student coffee_shots gender
<dbl> <dbl> <chr>
1 21 1 female
2 23 4 female
3 211 3 male
4 28 2 male - Add this column
genderto the joins
# A tibble: 187 × 6
subject condition gender mood_pre mood_post coffee_shots
<dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 2 control female 81 NA NA
2 1 stress female 59 42 NA
3 3 stress female 22 60 NA
4 4 stress female 53 68 NA
5 7 control female 48 NA NA
6 6 stress female 73 73 NA
7 5 control female NA NA NA
8 9 control male 100 NA NA
9 16 stress female 67 74 NA
10 13 stress female 30 68 NA
# ℹ 177 more rows- Exclude
gender, col names are kept and added suffixes -.xand.y
# A tibble: 187 × 7
gender.x gender.y coffee_shots subject condition mood_pre mood_post
<chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 female female 4 23 control 78 NA
2 male male 2 28 stress 53 68
3 female female 1 21 control 68 NA
4 female <NA> NA 2 control 81 NA
5 female <NA> NA 1 stress 59 42
6 female <NA> NA 3 stress 22 60
7 female <NA> NA 4 stress 53 68
8 female <NA> NA 7 control 48 NA
9 female <NA> NA 6 stress 73 73
10 female <NA> NA 5 control NA NA
# ℹ 177 more rowsTwo tables - same column names
Remark
Suffixes for columns with the same name can be controlled.
Join by one column
# A tibble: 187 × 7
gender_mood gender_coffee coffee_shots subject condition mood_pre mood_post
<chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl>
1 female female 4 23 control 78 NA
2 male male 2 28 stress 53 68
3 female female 1 21 control 68 NA
4 female <NA> NA 2 control 81 NA
5 female <NA> NA 1 stress 59 42
6 female <NA> NA 3 stress 22 60
7 female <NA> NA 4 stress 53 68
8 female <NA> NA 7 control 48 NA
9 female <NA> NA 6 stress 73 73
10 female <NA> NA 5 control NA NA
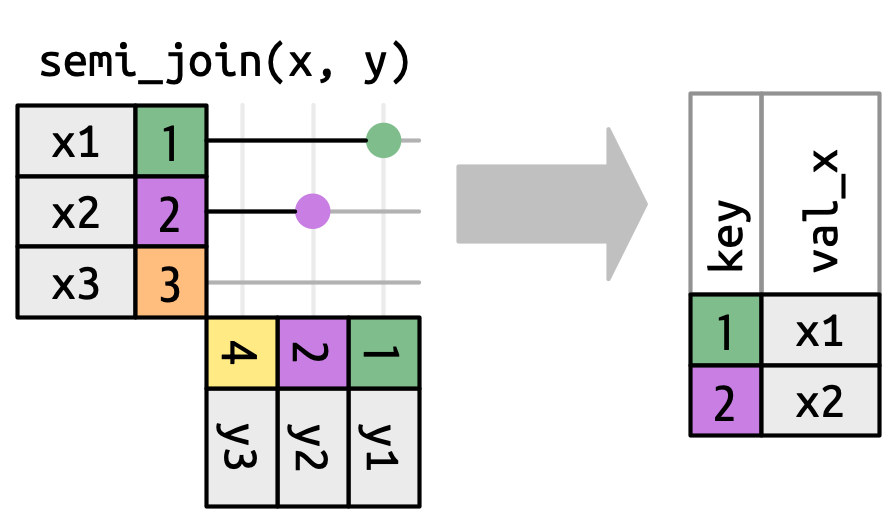
# ℹ 177 more rowsFiltering joins
Only the existence of a match is important; it doesn’t matter which observation is matched. This means that filtering joins never duplicate rows like mutating joins do
— Hadley Wickam
Filter matches in x, no duplicates
Extract what does not match
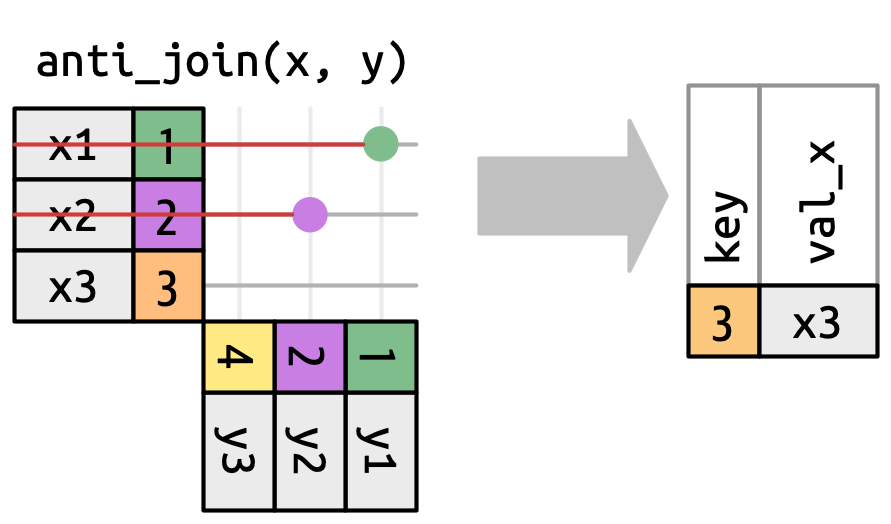
semi_join() does not alter original
# A tibble: 4 × 2
key x
<dbl> <chr>
1 1 x1
2 2 x2
3 3 x3
4 4 x4 When by = not precised, cols are reported (here, means key == key)
Cross join, Cartesian product
For permutations
# A tibble: 16 × 2
name.x name.y
<chr> <chr>
1 John John
2 John Simon
3 John Tracy
4 John Max
5 Simon John
6 Simon Simon
7 Simon Tracy
8 Simon Max
9 Tracy John
10 Tracy Simon
11 Tracy Tracy
12 Tracy Max
13 Max John
14 Max Simon
15 Max Tracy
16 Max Max Non-equi joins
Rolling joins
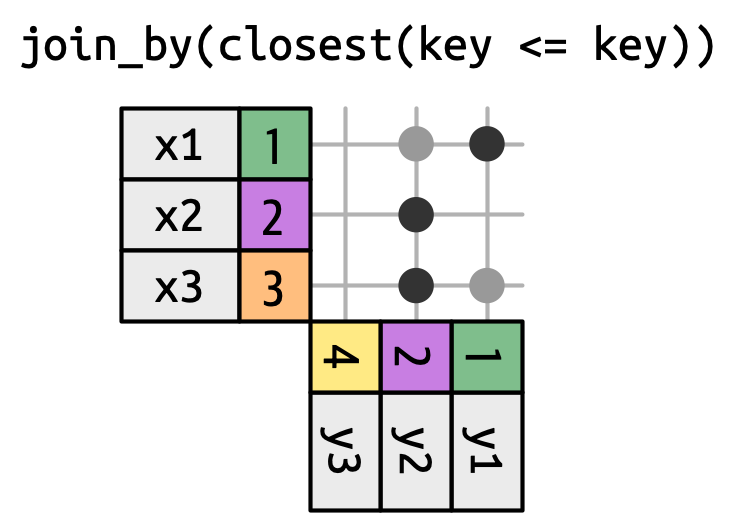
Not shown: Overlap joins with overlaps(), within() and between()
Helpful tools
Removing the data frame context through pull()
Remark
By default tidyverse operations that receive a tibble as input return a tibble.
Comparing to data in other rows
Leading and lagging rows
lead()for data in following rowlag()for data in the row above
Calculate differences between subjects
Let’s assume the subject IDs are in order of the tests being conducted. What is the difference to the previous subject for the “initial mood”?
# A tibble: 188 × 4
subject mood_pre prev_mood_pre mood_diff
<dbl> <dbl> <dbl> <dbl>
1 1 59 NA NA
2 2 81 59 22
3 3 22 81 -59
4 4 53 22 31
5 5 NA 53 NA
6 6 73 NA NA
7 7 48 73 -25
8 8 59 48 11
9 9 100 59 41
10 10 72 100 -28
# ℹ 178 more rowsThe other 20% of dplyr
Occasionally handy
- Assembly:
bind_rows,bind_cols - Windows function,
min_rank,dense_rank,cumsum. See - vignette - Working with list-columns (courses next year)
multidplyrfor parallelized code
SQL mapping allows database access
dplyrcode can be translated into SQL and query databases online (usingdbplyr)- Different types of tabular data (dplyr SQL backend, databases,
Before we stop
You learned to:
- Joining and intersecting tibbles
- Differ filtering from mutation joins
Acknowledgments 🙏 👏
- Hadley Wickham
- Lionel Henry
- Romain François
- Allison Horst for the great ArtWork
- Jenny Bryan
Contributions
- Milena Zizovic
- Roland Krause
Thank you for your attention!