# A tibble: 188 × 158
start_date end_date finished condition subject gender age mood_pre
<chr> <chr> <dbl> <chr> <dbl> <chr> <dbl> <dbl>
1 11/3/2014 11/3/2014 1 control 2 female 24 81
2 11/3/2014 11/3/2014 1 stress 1 female 19 59
3 11/3/2014 11/3/2014 1 stress 3 female 19 22
4 11/3/2014 11/3/2014 1 stress 4 female 22 53
5 11/3/2014 11/3/2014 1 control 7 female 22 48
6 11/3/2014 11/3/2014 1 stress 6 female 22 73
7 11/3/2014 11/3/2014 1 control 5 female 18 NA
8 11/3/2014 11/3/2014 1 control 9 male 20 100
9 11/3/2014 11/3/2014 1 stress 16 female 21 67
10 11/3/2014 11/3/2014 1 stress 13 female 19 30
# ℹ 178 more rows
# ℹ 150 more variables: mood_post <dbl>, STAI_pre_1_1 <dbl>,
# STAI_pre_1_2 <dbl>, STAI_pre_1_3 <dbl>, STAI_pre_1_4 <dbl>,
# STAI_pre_1_5 <dbl>, STAI_pre_1_6 <dbl>, STAI_pre_1_7 <dbl>,
# STAI_pre_2_1 <dbl>, STAI_pre_2_2 <dbl>, STAI_pre_2_3 <dbl>,
# STAI_pre_2_4 <dbl>, STAI_pre_2_5 <dbl>, STAI_pre_2_6 <dbl>,
# STAI_pre_2_7 <dbl>, STAI_pre_3_1 <dbl>, STAI_pre_3_2 <dbl>, …Grouping and summarizing
dplyr
University of Luxembourg
Saturday, the 25th of April, 2026
Grouping and summarizing
Reminder, we work with the judgments dataset:
Counting elements
How many participants per condition?
count()groups by specified columns- Result is a tibble that can be processed further
sort = TRUEavoid piping toarrange(desc(n))
Your turn!
- Count how many observations we have per
conditionandgender - Sort the tibble so the group with max observations is on the first row
- Bonus: the created column labelled
nobsinstead ofn(check the help page ofcount())
Solution
Summarise data
If we want the min/max per condition
# A tibble: 1 × 2
min max
<dbl> <dbl>
1 9 100summarisereturns as many rows as groups - one if no groups specified.mutatereturns as many rows as given.
# A tibble: 188 × 160
min max start_date end_date finished condition subject gender age
<dbl> <dbl> <chr> <chr> <dbl> <chr> <dbl> <chr> <dbl>
1 9 100 11/3/2014 11/3/2014 1 control 2 female 24
2 9 100 11/3/2014 11/3/2014 1 stress 1 female 19
3 9 100 11/3/2014 11/3/2014 1 stress 3 female 19
4 9 100 11/3/2014 11/3/2014 1 stress 4 female 22
5 9 100 11/3/2014 11/3/2014 1 control 7 female 22
6 9 100 11/3/2014 11/3/2014 1 stress 6 female 22
7 9 100 11/3/2014 11/3/2014 1 control 5 female 18
8 9 100 11/3/2014 11/3/2014 1 control 9 male 20
9 9 100 11/3/2014 11/3/2014 1 stress 16 female 21
10 9 100 11/3/2014 11/3/2014 1 stress 13 female 19
# ℹ 178 more rows
# ℹ 151 more variables: mood_pre <dbl>, mood_post <dbl>, STAI_pre_1_1 <dbl>,
# STAI_pre_1_2 <dbl>, STAI_pre_1_3 <dbl>, STAI_pre_1_4 <dbl>,
# STAI_pre_1_5 <dbl>, STAI_pre_1_6 <dbl>, STAI_pre_1_7 <dbl>,
# STAI_pre_2_1 <dbl>, STAI_pre_2_2 <dbl>, STAI_pre_2_3 <dbl>,
# STAI_pre_2_4 <dbl>, STAI_pre_2_5 <dbl>, STAI_pre_2_6 <dbl>,
# STAI_pre_2_7 <dbl>, STAI_pre_3_1 <dbl>, STAI_pre_3_2 <dbl>, ….before = or .after arguments to place wherever you want new columns
Summarise, but per specified goup: the .by argument
If we want the min/max per condition
# A tibble: 2 × 3
condition min max
<chr> <dbl> <dbl>
1 control 19 100
2 stress 9 96Works great in mutate() too:
Before v1.1.0, group_by() function (not recommended):
- No
tidyselecthelpers
# A tibble: 2 × 3
condition min max
<chr> <dbl> <dbl>
1 control 19 100
2 stress 9 96- Leave out grouped tibbles (here
condition)
# A tibble: 4 × 4
# Groups: condition [2]
condition gender min max
<chr> <chr> <dbl> <dbl>
1 control female 29 95
2 control male 19 100
3 stress female 9 96
4 stress male 18 85-Pipe to ungroup() to avoid that or groups = "drop". Or stick to .by!
Within one summarise statement
Commonly used
n()to count the number of rowsn_distinct()to count the number of distinct observations - used inside the dplyr verbs!first()to extract the observation in the first positionlast()to extract the observation in the last positionnth()to take the entry in a specified position
With a grouping variable or not
# A tibble: 1 × 5
n_rows n_subject first_id last_id id_10
<int> <int> <dbl> <dbl> <dbl>
1 188 187 2 189 13# A tibble: 2 × 6
gender n_rows n_subject first_id last_id id_10
<chr> <int> <int> <dbl> <dbl> <dbl>
1 female 147 147 2 189 18
2 male 41 40 9 182 48Dealing with multiple return values per group
range()returns min and maxsummarise()duplicates the key names
Error in `summarise()`:
ℹ In argument: `range = range(mood_pre, na.rm = TRUE)`.
ℹ In group 1: `condition = "control"`, `gender = "female"`.
Caused by error:
! `range` must be size 1, not 2.
ℹ To return more or less than 1 row per group, use `reframe()`.Use reframe() instead
# A tibble: 8 × 4
condition gender range n
<chr> <chr> <dbl> <int>
1 control female 29 65
2 control female 95 65
3 stress female 9 82
4 stress female 96 82
5 control male 19 26
6 control male 100 26
7 stress male 18 15
8 stress male 85 15Reframe with more values
mutate()requires the same number of rowssummarise()requires one value per groupreframe()accepts arbitrary number of rows per group and returns an ungrouped tibble.
New in dplyr 1.1.0
More advanced but useful: 3 quantiles
# A tibble: 12 × 5
condition gender q quartile n
<chr> <chr> <dbl> <dbl> <int>
1 control female 0.25 52.8 64
2 control female 0.5 66 64
3 control female 0.75 78.2 64
4 stress female 0.25 44 82
5 stress female 0.5 58.5 82
6 stress female 0.75 67 82
7 control male 0.25 53.2 26
8 control male 0.5 65 26
9 control male 0.75 72.8 26
10 stress male 0.25 45.5 15
11 stress male 0.5 53 15
12 stress male 0.75 69.5 15Your turn!
In judgments:
- Find the number of subjects by age, gender and condition, e.g. how many 20 years-old females are in the
controlgroup? - Sort the resulting tibble such that the condition that contains the most populous group is sorted first (i.e.
stressorcontrolappear together). - Ensure that the resulting tibble does not contain groups.
Solution
# A tibble: 33 × 4
condition gender age n
<chr> <chr> <dbl> <int>
1 control female 18 25
2 control female 19 17
3 control male 18 7
4 control female 21 7
5 control male 20 6
6 control male 22 5
7 control female 22 4
8 control female 20 4
9 control female 23 3
10 control male 23 3
11 control male 19 3
12 control female 24 2
13 control female 17 2
14 control female 26 1
15 control male 17 1
16 control male 21 1
17 stress female 18 27
18 stress female 19 19
19 stress female 20 14
20 stress female 22 9
21 stress female 17 5
22 stress female 21 4
23 stress male 18 4
24 stress male 22 3
25 stress male 19 3
# ℹ 8 more rowsAlternative:
# A tibble: 33 × 4
condition gender age n
<chr> <chr> <dbl> <int>
1 control female 18 25
2 control female 19 17
3 control female 21 7
4 control male 18 7
5 control male 20 6
6 control male 22 5
7 control female 20 4
8 control female 22 4
9 control female 23 3
10 control male 19 3
11 control male 23 3
12 control female 17 2
13 control female 24 2
14 control female 26 1
15 control male 17 1
16 control male 21 1
17 stress female 18 27
18 stress female 19 19
19 stress female 20 14
20 stress female 22 9
21 stress female 17 5
22 stress female 21 4
23 stress male 18 4
24 stress male 19 3
25 stress male 22 3
# ℹ 8 more rowsSummary
Most commonly used - 80%
select()- columnsfilter()- rows meeting conditionarrange()- sortglimpse()- inspect (not shown)rename()- change column namerelocate()- move columnsmutate()- create columnscase_when()simplifies if/else/if/elsesummarise()- group-wise summaries
Comments
- Go to the website for additional functions.
- R for datascience contains practical work through
across()powerful functional programming to act on multiple columns bonus slides.
Before we stop
You learned to:
- Grouping and summarizing
Next step: joining tables
Acknowledgments 🙏 👏
- Lise Vaudor nice blog
- Allison Horst for the great ArtWork
- Jenny Bryan
Contributions
- Milena Zizovic
- Roland Krause
Thank you for your attention!
Act on multiple columns with across()
Usage
Can be plugged into mutate(), summarise()…
across(ON WHO, DO WHAT)
- Columns selection:
- Argument
.cols tidyselecthelperseverything()= all columns.- Conditions (boolean) needs
where(),across(where(is.numeric))
- Argument
- Actions using functions:
- Argument
.fns fun, arg1, arg2\(x) fun(x), withplaceholderx- Multiple functions as arguments need to be wrapped up
- New column names can be controlled
- Argument
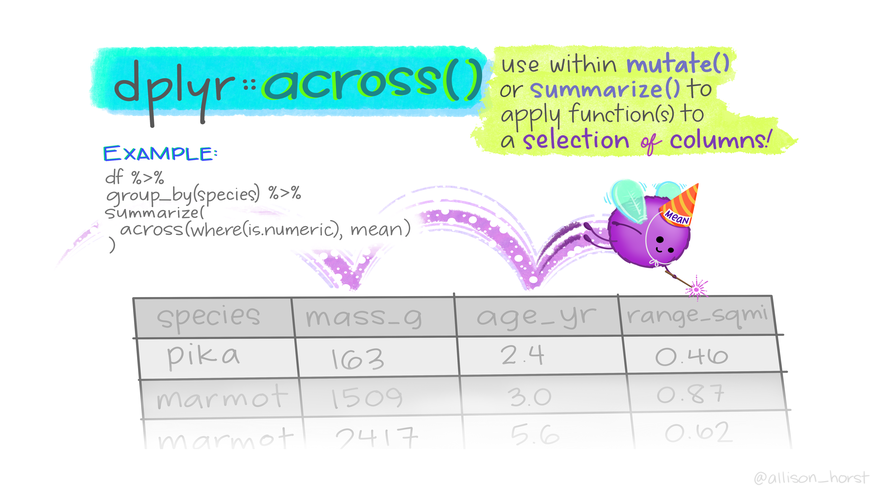
Artwork by Allison Horst
Examples of across() usage
Add 1 to all STAI columns
To convert Likert scales 0-4 to 1-5 (same column names)
# A tibble: 188 × 42
STAI_pre_1_1 STAI_pre_1_2 STAI_pre_1_3 STAI_pre_1_4 STAI_pre_1_5 STAI_pre_1_6
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 3 2 3 3 3 3
2 4 3 4 2 4 3
3 5 4 4 4 5 3
4 3 3 3 3 4 2
5 2 2 2 2 3 2
6 3 3 2 2 3 2
7 3 3 2 2 3 2
8 2 2 2 2 2 2
9 3 3 2 2 3 2
10 5 3 4 4 4 2
# ℹ 178 more rows
# ℹ 36 more variables: STAI_pre_1_7 <dbl>, STAI_pre_2_1 <dbl>,
# STAI_pre_2_2 <dbl>, STAI_pre_2_3 <dbl>, STAI_pre_2_4 <dbl>,
# STAI_pre_2_5 <dbl>, STAI_pre_2_6 <dbl>, STAI_pre_2_7 <dbl>,
# STAI_pre_3_1 <dbl>, STAI_pre_3_2 <dbl>, STAI_pre_3_3 <dbl>,
# STAI_pre_3_4 <dbl>, STAI_pre_3_5 <dbl>, STAI_pre_3_6 <dbl>,
# STAI_post_1_1 <dbl>, STAI_post_1_2 <dbl>, STAI_post_1_3 <dbl>, …Specify new names not to overwrite cols
# A tibble: 188 × 6
STAI_pre_1_1 STAI_pre_1_2 STAI_pre_1_3 incr_STAI_pre_1_1 incr_STAI_pre_1_2
<dbl> <dbl> <dbl> <dbl> <dbl>
1 2 1 2 3 2
2 3 2 3 4 3
3 4 3 3 5 4
4 2 2 2 3 3
5 1 1 1 2 2
6 2 2 1 3 3
7 2 2 1 3 3
8 1 1 1 2 2
9 2 2 1 3 3
10 4 2 3 5 3
# ℹ 178 more rows
# ℹ 1 more variable: incr_STAI_pre_1_3 <dbl>"STAI_pre_1_[1-3]" regular expression means "STAI_pre_1_" followed by either 1, 2, or 3
For filter, across is renamed to if any or all
Find rows where ANY of mood data columns are missing
Selecting columns with a predicate where()
Add 1 to all numeric columns
- Predicate means return
TRUEorFALSE
# A tibble: 188 × 158
start_date end_date finished condition subject gender age mood_pre
<chr> <chr> <dbl> <chr> <dbl> <chr> <dbl> <dbl>
1 11/3/2014 11/3/2014 2 control 3 female 25 82
2 11/3/2014 11/3/2014 2 stress 2 female 20 60
3 11/3/2014 11/3/2014 2 stress 4 female 20 23
4 11/3/2014 11/3/2014 2 stress 5 female 23 54
5 11/3/2014 11/3/2014 2 control 8 female 23 49
6 11/3/2014 11/3/2014 2 stress 7 female 23 74
7 11/3/2014 11/3/2014 2 control 6 female 19 NA
8 11/3/2014 11/3/2014 2 control 10 male 21 101
9 11/3/2014 11/3/2014 2 stress 17 female 22 68
10 11/3/2014 11/3/2014 2 stress 14 female 20 31
# ℹ 178 more rows
# ℹ 150 more variables: mood_post <dbl>, STAI_pre_1_1 <dbl>,
# STAI_pre_1_2 <dbl>, STAI_pre_1_3 <dbl>, STAI_pre_1_4 <dbl>,
# STAI_pre_1_5 <dbl>, STAI_pre_1_6 <dbl>, STAI_pre_1_7 <dbl>,
# STAI_pre_2_1 <dbl>, STAI_pre_2_2 <dbl>, STAI_pre_2_3 <dbl>,
# STAI_pre_2_4 <dbl>, STAI_pre_2_5 <dbl>, STAI_pre_2_6 <dbl>,
# STAI_pre_2_7 <dbl>, STAI_pre_3_1 <dbl>, STAI_pre_3_2 <dbl>, …More advanced across: multiple functions
Compute for mood cols
- means
- standard deviations
Better with cols renaming
# A tibble: 1 × 14
moral_dilemma_dog_aveg moral_dilemma_dog_sdev moral_dilemma_wallet_aveg
<dbl> <dbl> <dbl>
1 7.35 2.17 7.14
# ℹ 11 more variables: moral_dilemma_wallet_sdev <dbl>,
# moral_dilemma_plane_aveg <dbl>, moral_dilemma_plane_sdev <dbl>,
# moral_dilemma_resume_aveg <dbl>, moral_dilemma_resume_sdev <dbl>,
# moral_dilemma_kitten_aveg <dbl>, moral_dilemma_kitten_sdev <dbl>,
# moral_dilemma_trolley_aveg <dbl>, moral_dilemma_trolley_sdev <dbl>,
# moral_dilemma_control_aveg <dbl>, moral_dilemma_control_sdev <dbl>Your turn!
In judgments:
Compute basic statistics for all moral dilemma columns considering the condition (stress or control):
- Compute the mean, the median and the standard deviation.
- Find meaningful short names for the functions such as
medformedian(). - Column names should be prefixed by the short names from 2., not suffixes
- Assign the name
judgments_condition_statsto the results.
Solution
# A tibble: 2 × 22
condition mean_moral_dilemma_dog sd_moral_dilemma_dog med_moral_dilemma_dog
<chr> <dbl> <dbl> <dbl>
1 control 7.24 2.23 8
2 stress 7.45 2.11 8
# ℹ 18 more variables: mean_moral_dilemma_wallet <dbl>,
# sd_moral_dilemma_wallet <dbl>, med_moral_dilemma_wallet <dbl>,
# mean_moral_dilemma_plane <dbl>, sd_moral_dilemma_plane <dbl>,
# med_moral_dilemma_plane <dbl>, mean_moral_dilemma_resume <dbl>,
# sd_moral_dilemma_resume <dbl>, med_moral_dilemma_resume <dbl>,
# mean_moral_dilemma_kitten <dbl>, sd_moral_dilemma_kitten <dbl>,
# med_moral_dilemma_kitten <dbl>, mean_moral_dilemma_trolley <dbl>, …